HyperGen – Sustainability of Genomics Exploration Through Innovation
By Lisa Smith, Lead Research Data Engineer at the Wilmot Cancer Institute and Anna Brown, Software Architect at the Wilmot Cancer Institute
RNA-Sequencing
In 2020, John Ashton, Director of the Genomics Shared Resource for the Wilmot Cancer Institute, the University of Rochester Genomic Research Center, the GRC Bioinformatics, and the Functional Genomics Center, stated the importance of and the impending reliance on RNA sequencing. “Although analyses at the single-cell level have long been practiced in biology and medicine…the ability to perform whole transcriptome profiling [RNA sequencing (RNA-seq)] by next generation sequencing (NGS) at the resolution of single cells is a relatively recent and…very powerful approach.” Four years later, that approach is widely used because understanding the data provides clinicians with an increased understanding of the molecular basis of cancer and the complex biological processes involved in treating it [1].
At its core, RNA sequencing is a method of analysis for measuring the expression level of each gene in a strand of RNA. The data sets are extremely large and require processing pipelines and the use of analysis packages, often written in R, including BaySeq, edgeR, DESeq, and DESeq2. Each of these applications model gene expression levels, and the error inherent in that model differently; BaySeq uses a Bayesian analysis approach, while edgeR and DESeq2 use a combination of non-Bayesian (Poisson or Negative Binomial distribution) and Bayesian approaches to model the expression level and error. In 2022, Li, Zand, et al. performed a comprehensive analysis of the most used analysis methods and concluded that overall, while several of the applications worked well and DESeq2 is a top choice, there was a troubling lack of consistent results from the different methods and a profound need for new RNA-seq differential analysis methods [2].
Cost of Knowledge
While extremely valuable, the process of RNA sequencing, from analyses to understanding, is a time consuming and costly one. To develop new methods requires a huge amount of time – time to not only determine what is working best and where and how the current methods fall short, but to also architect solutions to address those shortcomings while preserving the current advantages.
At the University of Rochester GRC Bioinformatics Group (GRC), while the need for this information grew, time was increasingly scarce. To process and analyze RNA, the staff had created their own GitHub repository of R scripts to run on their own servers. Most of the processes and pipelines were run in the command line, and the associated analysis reports were painstakingly created from the output of those processes. For example, several output files would have to be “stitched” together to display the results from one run of DESeq2. Due to the complexity and number of steps involved, it wasn’t feasible for researchers to investigate the data autonomously. Instead, they increasingly submitted requests for staff at the GRC to run the processes and deliver the results. Additionally, because of the size of the data and the lack of automation, handling research requests used an immense amount of the limited server space and left little time for the GRC’s many other tasks, including almost no time for improvement or innovation.
Enter HyperGen
Since its debut, HyperGen has exceeded expectations and has delivered real sustainability to genomics research. Its self-service nature has allowed the GRC staff to reclaim a significant portion of their valuable time to put towards their own research and innovation, while still providing genomics insights to clinical researchers pushing the boundaries of cancer treatment.
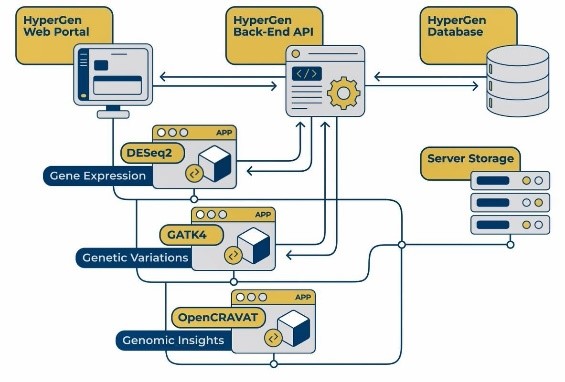
At the University of Rochester Medical Center’s Wilmot Cancer Institute, we have overcome many of the data-centric obstacles that typically plague the Healthcare setting by utilizing the Hyperion platform.[3] Hyperion allows researchers, admins, and clinicians to access and analyze real-time data in several user-friendly custom-built applications, including AI/ML, LLM, NLP, Blockchain, VR/AR, and Geospatial, so applying these approaches to genomics data was natural for us [4]. Working in collaboration with the GRC staff and applying the principles we have followed for the Hyperion suite of tools, we created HyperGen – a self-service genomics research discovery platform.
HyperGen puts genomics exploration back in the hands of researchers by enabling users to run RNA sequencing routines to explore genomics data, specifically DESeq2 to measure gene expression as well as other vital tools. HyperGen also provides users access to GATK4, to identify sample variants, and to perform advanced analysis via OpenCRAVAT, all from a single, secure web application. It is a completely custom platform built on open-source technologies in a containerized environment. HyperGen includes integrated routines for queueing jobs, reporting updates and errors to the users throughout the process, and delivering comprehensive analysis reports. The application also features a point-and-click, no-code interface with built-in tips and allows users to seamlessly log back in and view any recent results.
When using DESeq2 to analyze gene expression in HyperGen, users can upload normalized human or mouse count data, define the groupings for and order of comparison, title the run, and kick off the process. When the RNA sequencing is completed, the list of result files generated from DESeq2 and previews of some files are displayed for the user to view in the browser and download. Similarly, GATK4 can be used with the same features and user-friendly environment, to analyze gene variations. Lastly, HyperGen provides local, secure access to OpenCRAVAT as well, so researchers can further study their data without any need to leave the internal tool.
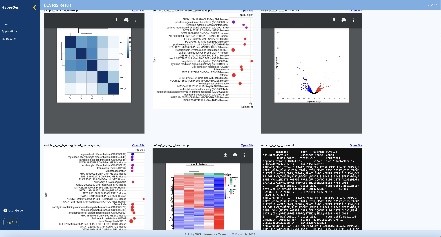
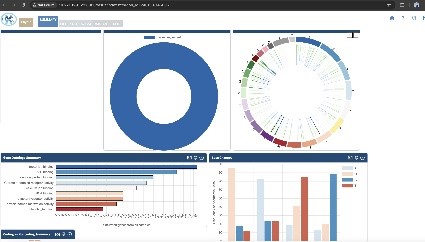
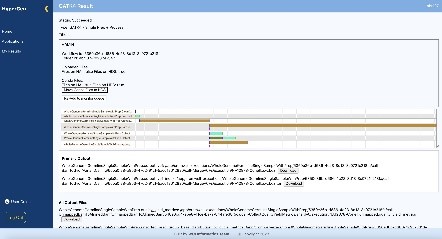
Sustainability
Since its debut, HyperGen has exceeded expectations and has delivered real sustainability to genomics research. Its self-service nature has allowed the GRC staff to reclaim a significant portion of their valuable time to put towards their own research and innovation, while still providing genomics insights to clinical researchers pushing the boundaries of cancer treatment. Its user-friendly, intuitive interface encourages exploration by providing seamless transitions between and within tools and a history of completed investigations. Powered by the Hyperion philosophy and architecture, and built completely in house, HyperGen requires no vendor contracts, licenses, or ongoing fees of any kind. It is a sustainable innovation supporting sustainable innovation in the fight to cure cancer.
Resources
1 Ashton JM, Rehrauer H, Myers J, et al. Comparative Analysis of Single-Cell RNA Sequencing Platforms and Methods. J Biomol Tech. 2021 Dec 15;32(4):3fc1f5fe.3eccea01. doi: 10.7171/3fc1f5fe.3eccea01. PMID: 35837267; PMCID: PMC9258609.
2 Li D, Zand MS, Dye TD, et al. An evaluation of RNA-seq differential analysis methods. PLoS ONE. 2022 Sept 16; 17(9): e0264246. https://doi.org/10.1371/journal.pone.0264246
3 Snyder E, Rivers T, Smith L, et al.From months to minutes: creating Hyperion, a novel data management system expediting data insights for oncology research and patient care. PLOS Digital Health (2022) https://doi.org/10.1101/2022.04.06.22273493
4 Smith L, Snyder E Geospatial Analysis: Changing the Landscape of Healthcare Informatics by Moving Beyond Typical Business Intelligence Tools, CXOTech Magazine, 06/2023 https://cxotechmagazine.com/geospatial-analysis-changing-the-landscape-of-healthcare-informatics-by-moving-beyond-typical-business-intelligence-tools/

